Global Analysis of Human Signaling Networks
To obtain a comprehensive understanding of the molecular mechanisms that regulate the cellular machinery, we are systematically examining families of regulatory molecules by undertaking some large-scale studies to complement our pathway-focused approaches.
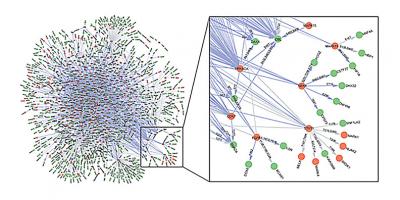
A high-resolution phosphorylation network: In collaboration with Heng Zhu and Jiang Qian’s laboratories at Johns Hopkins, we developed a new strategy, called CEASAR, based on functional protein microarrays and bioinformatics to experimentally identify substrates for 289 unique kinases, resulting in 3656 high-quality KSRs. We then generated consensus phosphorylation motifs for each of the kinases and integrated this information, along with information about in vivo phosphorylation sites determined by MS, to construct a high-resolution map of phosphorylation networks that connects 230 kinases to 2591 in vivo phosphorylation sites in 652 substrates.