We are interested in understanding how cells, the basic units of life, sense changing environments and orchestrate specific responses to carry out life processes. Recent years have seen tremendous progress in identifying the molecular components that constitute the structural, biochemical and mechanical networks that control various life processes. Less well developed is our understanding of how these components are precisely regulated to achieve functional specificity within a living cell, which may be simultaneously reacting to multiple inputs from the changing environment. We believe the key lies in the spatiotemporal information encoded in a particular cellular context.
We are investigating the molecular mechanisms and functional roles of such spatiotemporal regulation by taking a “native biochemistry” approach. This allows us to unravel the chemical mechanisms of life processes in the framework of cellular time and space, where molecular changes can be directly linked to functional effects. Work in the Zhang laboratory combines genetically encoded fluorescent biosensors, superresolution imaging, targeted biochemical perturbations and mathematic modeling to investigate the spatiotemporal regulation of the cAMP/PKA, Ca 2+ /calcineurin, PI3K/Akt/mTOR, MAPK and AMPK pathways in the context of energy metabolism, axon polarization and genesis, insulin secretion by β cells, as well as tumorigenesis.
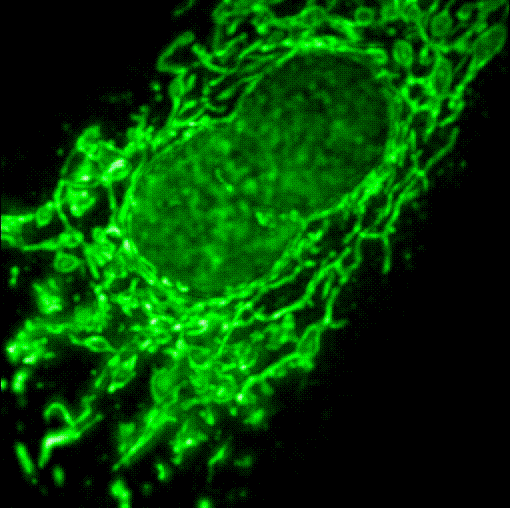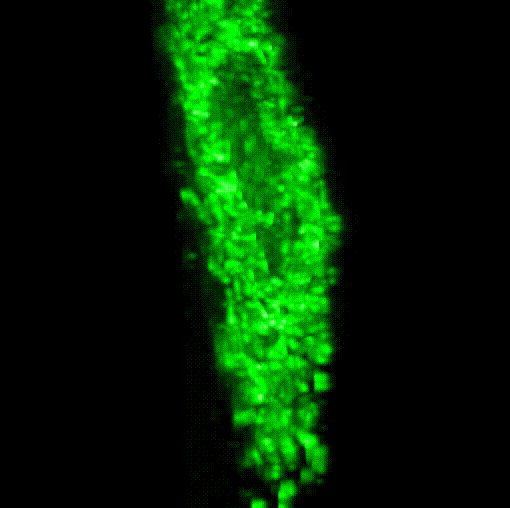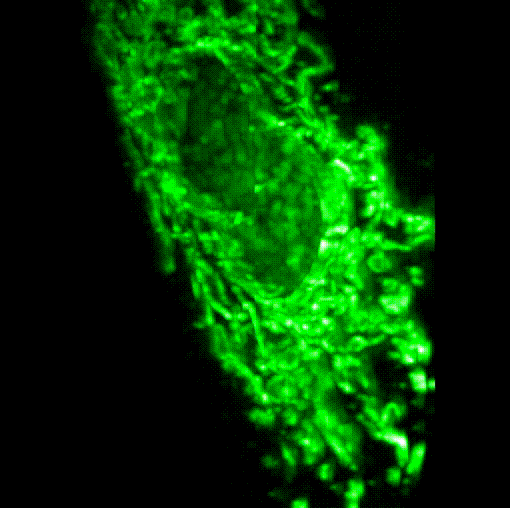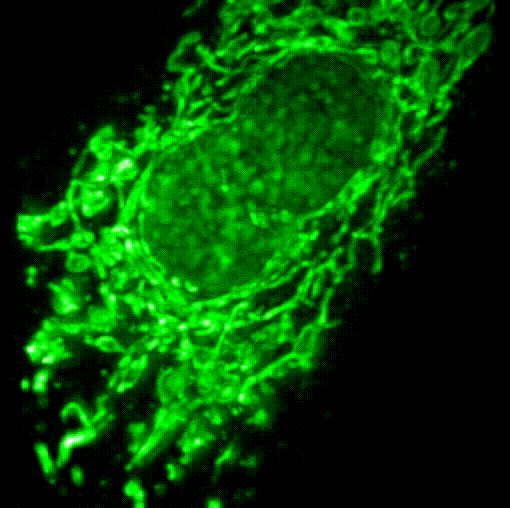
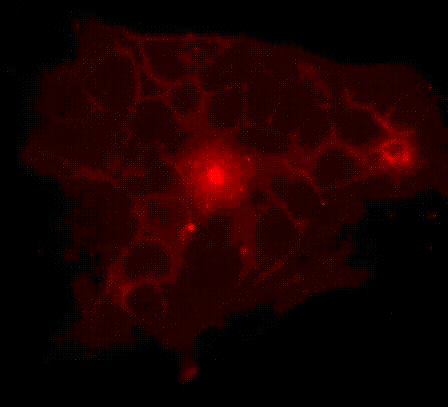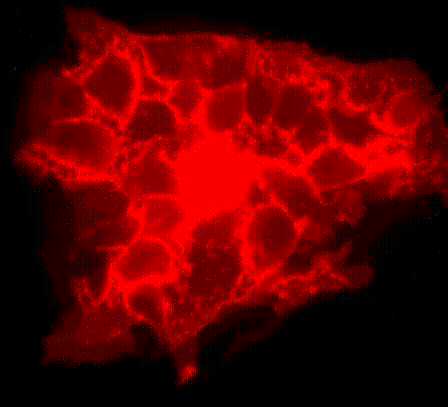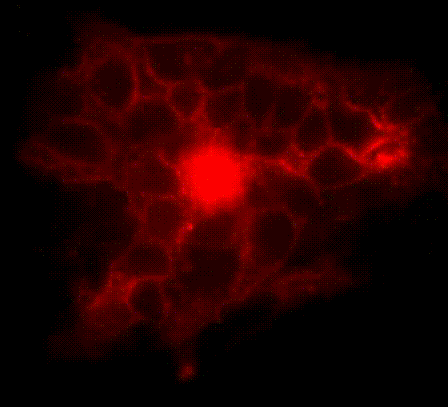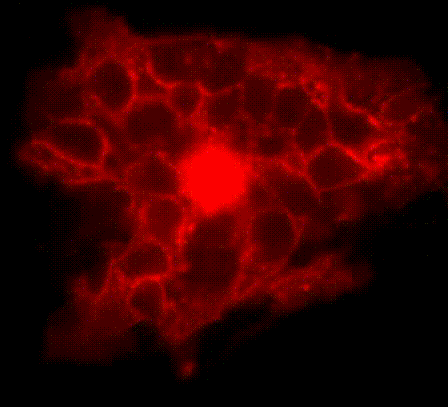
We have developed a series of molecular tools and fluorescence imaging technologies to interrogate signaling enzymes in their native contexts. For instance, general strategies have been developed for visualizing signaling activities in living cells, leading to the creation of a series of activity biosensors for various protein kinases, phosphatases and second messengers. We are currently expanding our biosensor toolkit, engineering novel magnetic labels and developing strategies for obtaining super-resolution maps for signaling activities.